Tb ADAT2/ADAT3 deaminase in complex with tRNA
* ADAT : Adenosine Deaminase Acting on tRNA
Nature Communications 2022 Nov 8;13(1):6737.
The Cryo-EM structure 8AW3 [3D viewer LINK]
Abstract
The essential deamination of adenosine A34 to inosine at the wobble base is the individual tRNA modification with the greatest effects on mRNA decoding, empowering a single tRNA to translate three different codons. To date, many aspects of how eukaryotic deaminases specifically select their multiple substrates remain unclear. Here, using cryo-EM, we present the structure of a eukaryotic ADAT2/3 deaminase bound to a full-length tRNA, revealing that the enzyme distorts the anticodon loop, but in contrast to the bacterial enzymes, selects its substrate via sequence-independent contacts of eukaryote-acquired flexible or intrinsically unfolded motifs distal from the conserved catalytic core. A gating mechanism for substrate entry to the active site is identified. Our multi-step tRNA recognition model yields insights into how RNA editing by A34 deamination evolved, shaped the genetic code, and directly impacts the eukaryotic proteome.
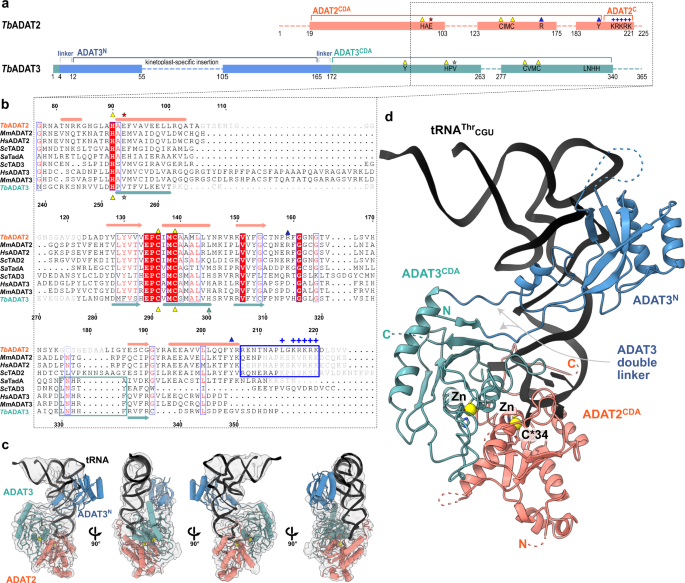
a Schematic representation of the protein subunits: ADAT2 CDA in salmon, ADAT3 N-terminal domain in blue, ADAT3CDA in teal. Portions with traced atomic model as solid colors and non-resolved linkers as dotted lines. Important sequence motifs are annotated at their relative positions. Zn2+ coordinating residues are indicated by yellow triangles, the active-site glutamate / pseudo-active-site valine by a red and gray star, ‘gate’ residues of ADAT2 with a blue triangle. Conserved positive charges of the KR-motif annotated as blue “+“ signs. b Structural alignment of Tb, yeast and mouse ADAT2 and ADAT3 CDA domains and Staphylococcus aureus TadA. The sequences for human (Hs) ADAT2/3 included (MmADAT2/3 PDB: 7NZ7, scTAD2/3 PDB:7BV5, saTadA PDB: 2B3J). Structural elements of Tb are annotated (ADAT2 above, ADAT3 below) with α-helices as cylinders and β-sheets as arrows. Zn2+ coordinating residues are indicated by yellow triangles, the active-site glutamate / pseudo-active-site valine by a red and gray star, ‘gate’ residues of ADAT2 with a blue triangle and the active-site histidine of ADAT3 with a teal triangle. ADAT2C is boxed in blue with annotation of the conserved positive charges of the KR-motif (“+“ signs). Grayed out letters indicate non-resolved loops in our structure and ADAT2C residues not resolved in previous crystal structures. An ADAT3 conserved loop is annotated as a teal box. Other colors and boxes within the alignment represent relative conservation above 70%. c Four views of the final non-b-factor-sharpened cryoSPARC map that was used for modeling (presented at a level of 0.09) with atomic model. Colors as above. d Cartoon model of ADAT2/3 bound to tRNA. Colors as in 1 A. Zn2+ atoms in yellow, tRNA in black with only anticodon loop bases represented as slabs, the reactive C34 nucleotide is annotated as C*. Gray arrows point to the ADAT3 N-terminal domain double linker.
The anticodon loop of the tRNA is remodeled upon ADAT2/3 binding
We inspected the overall conformation of the tRNA in the complex and noticed that the conformation of the anticodon-stem loop bound to the TbADAT2/3 core dramatically deviates from canonical free or ribosome A-site bound tRNA structures (Fig. 2a). In our complex structure, the TbtRNAThrCGU is deeply inserted into the active site of TbADAT2 with all seven nucleotides (32–38) of the anticodon loop interacting directly with the ADAT2/3 deaminase core (Fig. 2b). While the stem nucleotides strictly obey Watson-Crick base pairing, and the non-canonical base transition pair between C32 and A38 is maintained46, the loop nucleotides 33–37 are extensively remodeled. Here, the sugar-phosphate backbone adopts a bent conformation and the nucleosides U33, C*34, G35 and A37 are splayed outwards (Fig. 2c and Supplementary Fig. 3a). The nucleobase of U36 is unconventionally internalized via a π-π stacking interaction between the nucleobase of C32 and residue Y205 of ADAT2 (Fig. 2d and Supplementary Fig. 3a). The unusual ADAT2/3-bound anticodon loop conformation with exposed bases, similar to the TadA bound hairpin, was unexpected, because the eukaryotic deaminase does not rely on sequence read-out24,37,41.
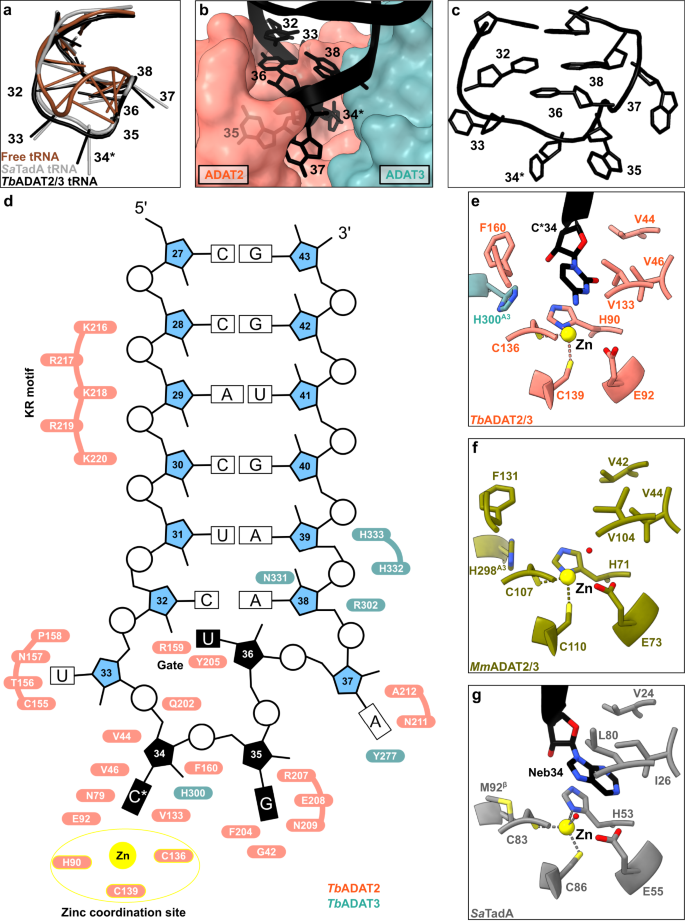
a The anticodon loop (ACL) bound to TbADAT2/3 (black) is distorted with respect to free tRNA (brown; PDB: 1EHZ) but resembles the ACL bound to bacterial TadA (gray; PDB: 2B3J), all cartoon-stick representation. b A deep cleft in the dimer interface accommodates the ACL. Colors as in Fig. 1, ADAT2/3 model as surface representation, tRNA as cartoon with anticodon loop bases shown. c The deformed ACL bound to the TbADAT2/3, backbone in cartoon and bases in stick representation. d Schematic overview of the anticodon stem loop (ACSL) and its interactions with TbADAT2/3. Zinc coordinating site highlighted in yellow. e–g Comparison of the active-site pockets of Tb and MmADAT2/3 and TadA in the same orientation: e TbADAT2/3 with tRNA cytosine-base shown; f apo-MmADAT2/3 (PDB: 7NZ7), with the conserved H298MmADAT3 as the only ADAT3 residue present in the active site; g bacterial SaTadA (PDB: 2B3J) with nebularine bound, with M92SaTadA훃 residue contributed by the second protomer of the homodimer. Residue side chains colored by heteroatom with zinc atoms in yellow.
A deep catalytic pocket in ADAT2/3 accommodates A34
To determine the molecular basis for the unusual rearrangement of the tRNA, we examined how the binding pocket accommodates the anticodon loop. This pocket lies in the dimer interface between the cytidine deaminase (CDA) domains of TbADAT2 and TbADAT3. The flipped-out A34 (C34 in our construct) is deeply inserted into the active site of TbADAT2 (Fig. 2b). The active site of TbADAT2 features the typical CDA elements: a Zn2+ cation coordinated by two cysteines (C136TbADAT2 and C139TbADAT2) and a histidine (H90TbADAT2), and the catalytic glutamate (E92TbADAT2), suggesting a similar catalytic mechanism as described for TadA24. Furthermore, the reactive nucleobase C*34 conformation is stabilized through hydrophobic interactions with V46TbADAT2 and V133TbADAT2 as well as hydrogen bonding with the conserved asparagine N79TbADAT2, while its ribose moiety is clamped by V44TbADAT2 and F160TbADAT2 (Fig. 2e and Supplementary Fig. 3b). The catalytic glutamic acid side chain, E92TbADAT2, and the catalytic water could not fully be resolved in our maps (Supplementary Fig. 3b). Besides these observations, the binding pocket for nucleotide A34 is remarkably similar between the tRNA-bound and -unbound structures, pointing to a certain rigidity of the active site to which the tRNA ligand must adapt (compare Fig. 2e, f, and Supplementary Fig. 3c). Remarkably, most residues in direct proximity to A34 are conserved between the bacterial and eukaryotic enzymes (Fig. 2g). An exception is the presence of histidine H300TbADAT3, which is conserved within ADAT3s (Fig. 1b, compare Figs. 2e, g). In summary, the central ADAT2/3 catalytic pocket for the C*34 nucleotide base is highly conserved between the bacterial homodimer and the eukaryotic heterodimer and no large conformational changes can be observed between the available eukaryotic substrate-bound and -unbound A34 pockets. Taken together these observations imply that for binding, the tRNA must adapt to ADAT2/3 through structural rearrangements.
The anticodon loop is anchored by a molecular gate in ADAT2
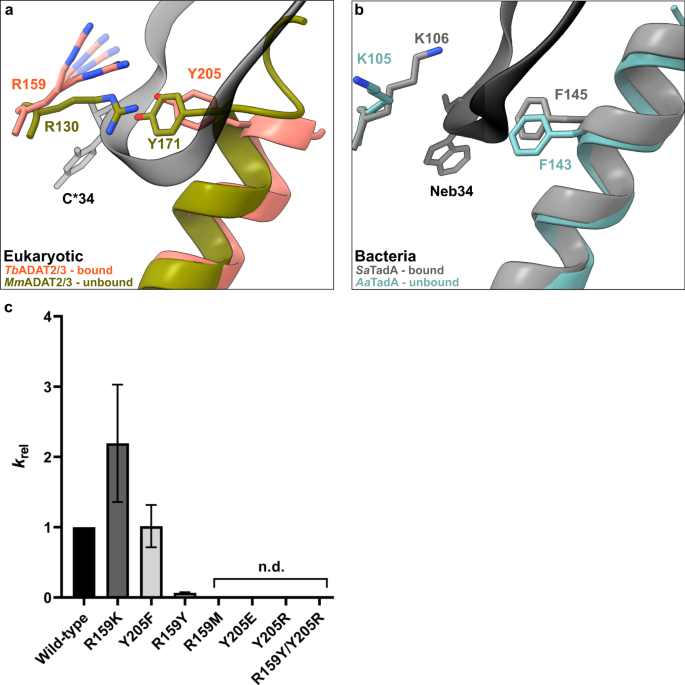
In the bacterial enzyme TadA, extensive remodeling of the anticodon loop is provoked by favorable base-specific interactions, resulting in splayed-out anticodon loop nucleosides bound to a rather rigid protein surface24. Since base-specific recognition does not take place in ADAT2/337,41, we looked for other features that could evoke such a non-canonical anticodon loop rearrangement. We identified a eukaryote-specific molecular “RY-gate” formed by residues arginine R159TbADAT2 and tyrosine Y205TbADAT2, which may provide a certain control mechanism for the tRNA to overcome before fully entering the catalytic pocket (Fig. 3). In the RNA-free MmADAT2/3 structures (PDB: 7NZ7, 7NZ8), this gate is closed through a cation-π interaction between these residues, R130MmADAT2 and Y171MmADAT2 (Fig. 3a and Supplementary Fig. 3f). In the tRNA-bound TbADAT2/3 structure, the gate arginine R159TbADAT2 is released, and appears to be flexible as judged by the map density (Supplementary Fig. 3d). Opening of this gate also frees the side chain of the second gate residue Y205TbADAT2, which together with nucleobase C32, can sandwich and internalize nucleotide U36 via π-π stacking. This is likely to be a major driver in remodeling the anticodon loop (Supplementary Fig. 3e). Interestingly, despite the presence of an equivalent positively charged-aromatic pair (e.g., K106SaTadA and F145SaTadA), this bacterial “gate” does not seem to operate because of suboptimal interacting geometry, which maintains these residues in an open conformation regardless of the presence or absence of RNA substrate; all available bacterial TadA structures in the PDB to date show an open configuration of these residues (Fig. 3b)21,24,47,48. In an earlier study, the substitution of R159TbADAT2 to alanine displayed unaltered RNA-binding properties39, so next we set out to scrutinize the importance of the gate residues in catalysis through deamination assays. All amino acid substitutions that removed the aromatic nature of Y205TbADAT2 (namely Y205E and Y205R) had no detectable activity after 24 h incubation, while a conservative mutation that maintains the aromaticity (Y205F) showed a reaction rate comparable to the wild-type enzyme. The necessity of the aromatic residue in this position indicates its importance in the base-sandwich for anticodon loop distortion. Next, we substituted the positive charge of the cationic gate residue R159TbADAT2 with a hydrophobic methionine (R159M), which led to an inactive enzyme. However, replacing R159TbADAT2 with tyrosine did not inactivate the enzyme but led to a 10-fold reduction of activity, indicating the importance of a positive charge for this gate residue. An “inverted gate” double mutant (R159Y, Y205R) was also inactive. A mutant mimicking the bacterial situation with a R159K substitution, conserving the positive charge but altering its geometry, strikingly, showed a more than 2-fold improved reaction rate compared to the wild-type, suggesting a lysine in this position is more efficient for deamination activity (Fig. 3c, d, Supplementary Fig. 3j, k, Supplementary Table 2). Once it has entered the gate, the anticodon loop gains access to the deep joint anticodon binding pocket in order to establish further interactions with the ADAT2 core: asparagine N157TbADAT2 can form a hydrogen bond with the hydroxyl of the U33 ribose and the splayed-out nucleotide U33 can interact with ADAT2 residues 155TbADAT2−158TbADAT2 as well as glutamine Q202TbADAT2 (Fig. 2d). An ADAT3 RNA-binding loop, 328–333TbADAT3, conserved between TadA and ADAT3 but not ADAT2, establishes contacts with the tRNA backbone at nucleotides 38–40 and the nucleobase A38 via two histidines (H332TbADAT3 and H333TbADAT3) and an asparagine N331TbADAT3, respectively (framed teal in Fig. 1b, Fig. 2d, Supplementary Fig. 3g–i). In conclusion, we identify a molecular gate formed by a cation-π interaction in eukaryotic ADAT2 which needs to be penetrated by the tRNA; the opened gate allows stabilizing aromatic stacking interactions with the C32 and U36 nucleobases to facilitate access to the deep anticodon loop binding cleft where further favorable protein–RNA interactions are established, altogether promoting the unusual anticodon loop configuration.
a Superposition of the tRNA-bound gate in TbADAT2/3 and the apo-gate in MmADAT2/3 (PDB: 7NZ7). The side chain of R159TbADAT2 is represented as flexible due to its poor side chain density. b The corresponding positions in bacteria deaminase TadA, RNA bound versus unbound (PDB: 2B3J and 1WWR, respectively). c Fold change of the observed rate constants of mutants to wild-type (krel = kobs mutant/kobs WT). Graph values calculated from three independent replicates. All mutants are of TbADAT2 co-expressed with TbADAT3. n.d. denotes no detectable activity after 24 h assay incubation. kobs values obtained from single-turnover kinetic assays (n≥3). Source data are provided as a Source Data file.
The ADAT2 C-terminus embraces the anticodon-stem loop
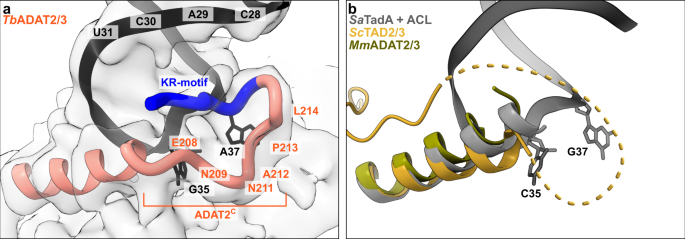
The available crystal structures of apo-ADAT2/3 failed to resolve the very C-terminal portion of ADAT2 (ADAT2C, residues 207–225TbADAT2), a segment which is absent in bacterial TadA proteins. This region carries a conserved, positively charged motif at the C-terminal end of ADAT2C, herein referred to as “KR-motif” (residues 216–220TbADAT2 KRKRK) (Fig. 1b). The KR-motif was previously identified as essential for tRNA binding and deamination39. We fitted ADAT2C into the cryo-EM map guided by the AlphaFold model of the complex; following the C-terminal helix which is the last element resolved in the crystal structures, a conserved proline induces a turn of the polypeptide, such that the KR-motif bends back towards the helix (Fig. 4, Supplementary Fig. 1g). ADAT2C displays extensive contacts with the anticodon loop and stem: residues 207–209TbADAT2 and 210–213TbADAT2 of TbADAT2C embrace the flipped-out nucleobases G35 and A37 (Fig. 2d). Furthermore, the positively charged C-terminal KR-motif, which aligns into the major groove of the anticodon loop, likely provides extended interactions to the phosphate backbone of the anticodon stem towards nucleotides 28–31 (Fig. 4a). The less clear density here might reflect a certain fluidity of the interaction and adaptation to different sequence registers could be imagined, which would be required to adjust to the different types of tRNA. Taken together, ADAT2C including the essential KR-motif seems to be intrinsically flexible in the absence of tRNA, but can reorganize and adjust to bind into the major groove of the anticodon loop and also establish interactions with portions of the anticodon stem.
a The C-terminus of ADAT2 embraces nucleotides 35 and 37 and KR-loop residues (K or R) provide additional contacts with the anticodon-stem loop via phosphate backbone interactions at residues 28–31. A cartoon representation of the ADAT2 C-terminus and tRNA is shown with the final non-b-factor-sharpened cryoSPARC map. Due to the limited resolution, we refrain from showing side chains in the ADAT2C model. b The same region of ScTAD2/3 (PDB: 7BV5), MmADAT2/3 (PDB: 7NZ7), and RNA bound SaTadA (PDB: 2B3J). The flexible unmodeled loop of ScADAT2 is shown as a dotted line.
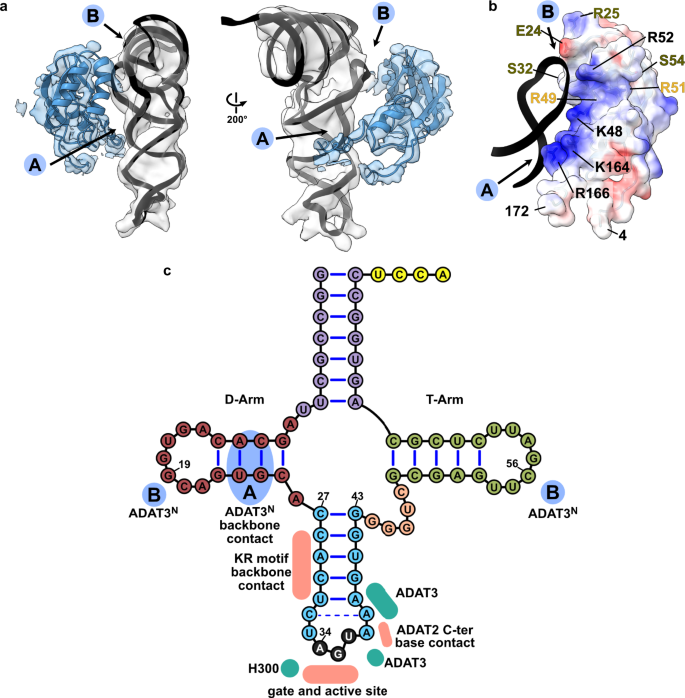
a Two views of the ADAT3N fitted in the post-processed EM map, in blue, with ‘A’ and ‘B’ regions as the two contact points of our model with tRNA, as indicated in C. b ADAT3N including linkers (residues 4–172); surface model colored by vacuum electrostatic potential, positive charges in red and negative charges in blue, with D-Arm of tRNA in a black cartoon representation. Residues identified to play a role in tRNA binding are annotated in black, in olive from41 and in yellow from40. (c) Schematic of the tRNA with interaction contacts. Anticodon-stem loop interactions are represented as a summary of the ones indicated in Fig. 2D. The two ADAT3N contact points, ‘A’ and ‘B’ are indicated in blue as above.
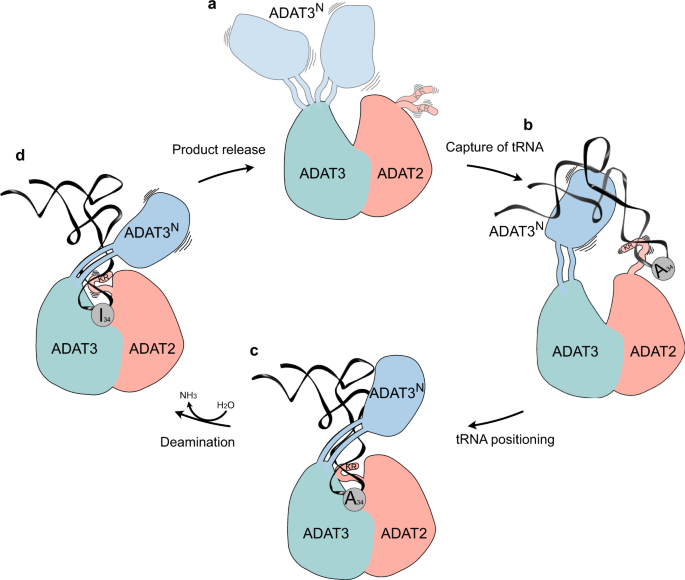
a Ligand-free enzyme state: in the free ADAT2/3 heterodimer ADAT2C is disordered and ADAT3N is flexibly linked. b Initial capture: the RNA recognition motifs in ADAT3N and in ADAT2C capture RNA; tRNAs are progressively guided by the extension of favorable interactions towards the anticodon binding cleft; hereby non-tRNA architectures will be rejected. c Anticodon loop insertion into the active site: The anticodon loop enters the active-site cleft through the RY-gate, and ADAT2C folds into the major groove of the anticodon loop to position the ligand for the reaction. d Substrate release: Detachment of the individual RNA-binding motifs initiates tRNA release.
Acknowledgements
We acknowledge Martin Pelosse for support in using the Eukaryotic Expression Facility at EMBL Grenoble, and Sarah Schneider for support in using the EM Facility at EMBL Grenoble. This work benefited from access to the cryo-EM platform of the Structural and Computational Biology Unit at EMBL Heidelberg. We acknowledge the HTX Team for the thermostability assay. We acknowledge the EMBL Genomic Core Facility for the tRNA sequencing. This work used the platforms of the Grenoble Instruct-ERIC center (ISBG; UAR 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology (PSB), supported by FRISBI (ANR-10-INBS-0005-02) and GRAL, financed within the University Grenoble Alpes graduate school (Écoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003). We thank Aline Le Roy and Christine Ebel, for assistance and access to the Protein Analysis On Line (PAOL) platform. We thank the Kowalinski lab members for discussion and comments throughout the course of the project. We also thank Andrew McCarthy and Sebastian Falk for comments on the manuscript. The present work was funded in part by the National Institutes of Health Grant GM084065-11 to J.D.A. The work was supported by a grant from the French Agence Nationale de la Recherche to E.K. (ANR-20-CE11-0016).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dolce, L.G., Zimmer, A.A., Tengo, L. et al. Structural basis for sequence-independent substrate selection by eukaryotic wobble base tRNA deaminase ADAT2/3. Nat Commun 13, 6737 (2022). https://doi.org/10.1038/s41467-022-34441-z
Received
Accepted
Published
DOIhttps://doi.org/10.1038/s41467-022-34441-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.